INNOVATION OF 2019| TOP 10 INNOVATION
INNOVATION OF 2019| TOP 10 INNOVATION
 |
INNOVATION OF 2019| TOP 10 INNOVATION
This year has proven to be a great one for the life sciences, and the 2010s have been an exciting decade. Here, The Scientist presents the fine-tuned tweaks and brand-new technologies that make up our Top 10 Innovations for 2019.
Refeyn Refeyn OneMP  In 2018, Philipp Kukura at the University of Oxford and his colleagues announced a technology they had developed, called mass photometry, that measures the weight of single molecules by the way they scatter light. The approach offered an entirely novel way to analyze biomolecules, with a few key advantages. Instead of moving samples into a vacuum as native mass spectrometry requires, mass photometry allows them to stay in their buffer solution. And unlike chromatography, which requires rather large sample volumes, mass photometry can read just a few microliters of a nanomolar concentration solution. To top it all off, the processing time is about a minute or two, compared to an hour or so for chromatography. In 2018, Philipp Kukura at the University of Oxford and his colleagues announced a technology they had developed, called mass photometry, that measures the weight of single molecules by the way they scatter light. The approach offered an entirely novel way to analyze biomolecules, with a few key advantages. Instead of moving samples into a vacuum as native mass spectrometry requires, mass photometry allows them to stay in their buffer solution. And unlike chromatography, which requires rather large sample volumes, mass photometry can read just a few microliters of a nanomolar concentration solution. To top it all off, the processing time is about a minute or two, compared to an hour or so for chromatography.
In short order, the University of Oxford spun out Refeyn to commercialize mass photometry, and its Refeyn OneMP instruments went on the market in March of 2019, starting at roughly $150,000. Matthias Langhorst, the chief marketing officer of the company, says applications for the tool range from simple tests of purity—the number of mass peaks will indicate how many types of protein are in a sample, for example—to more sophisticated analyses of how biomolecule assemblies behave under different conditions.
Thomas Schwartz, who studies the cell’s nuclear pore complex at MIT, was an early tester of the device, reaching out to Kukura as soon as he heard about mass photometry. “The most valuable thing with this instrument is that we can look at complex proteins-macromolecular complexes and figure out what are the components in that mixture, and do we detect any issue with stability,” he says. “It’s a very time-consuming process in typical workflow.”
Wiley: “The principles are not new, but the implementation in this new device is quite clever and powerful because it can measure every particle in a field. Very powerful and potentially revolutionary instrument.”
NeuBase Janus bases  Researchers can silence RNA by designing antisense oligonucleotides that complement the target sequence; the first therapeutics based on this approach are just hitting the market. (See “Oligonucleotide Therapeutics Near Approval,” The Scientist, December 2016.) But manufacturing long oligonucleotides and delivering them to cells are not easy tasks, says Carnegie Mellon synthetic organic chemist Danith Ly. Researchers can also target RNA with small molecules, which are easier to manufacture and deliver but don’t selectively bind to the targeted RNA. “We thought we might be able to find a sweet spot between the two traditional methods,” says Ly, who with his former postdoc Shivaji Thadke developed double-sided Janus bases (Comm Chem, 1:79, 2018). Researchers can silence RNA by designing antisense oligonucleotides that complement the target sequence; the first therapeutics based on this approach are just hitting the market. (See “Oligonucleotide Therapeutics Near Approval,” The Scientist, December 2016.) But manufacturing long oligonucleotides and delivering them to cells are not easy tasks, says Carnegie Mellon synthetic organic chemist Danith Ly. Researchers can also target RNA with small molecules, which are easier to manufacture and deliver but don’t selectively bind to the targeted RNA. “We thought we might be able to find a sweet spot between the two traditional methods,” says Ly, who with his former postdoc Shivaji Thadke developed double-sided Janus bases (Comm Chem, 1:79, 2018).
These bases are covalently linked to a charge-neutral peptide nucleic acid backbone, rather than a negatively charged sugar backbone, so the molecules can get cozy with target RNA or DNA. Janus bases also have two binding faces, allowing them to slip between the two sides of double-stranded nucleic acids, and bind to both of the complementary strands with stronger affinity than the two natural strands have to each other. Janus base therapeutics can therefore be highly specific to their targets even at lengths much shorter than traditional oligonucleotides.
NeuBase Therapeutics, founded in 2018 to develop peptide nucleic acid (PNA) therapeutics, exclusively licensed Janus bases from Carnegie Mellon last year and took Thadke on as its director of chemistry. The company now has two Janus-based drugs in early development, for Huntington’s disease and for myotonic dystrophy type 1. Each drug is just three Janus bases long and binds to two sides of an aberrant hairpin that forms in mutant RNAs that underlie disease pathogenesis. The company is now beginning mouse experiments, says Dietrich Stephan, cofounder, chairman, and CEO of NeuBase, with clinical trials projected to begin in 2021. He adds, “We’ve got a pipeline of other targets below that.”
Kamdar: “Development of a new gene modifying therapy with ability to target neurological and neuromuscular disorders. Combines the advantages of small molecules with the selectivity of antisense oligonucleotides.”
Berkeley Lights Lightning optofluidic platform  Berkeley Lights’s Lightning optofluidic platform enables researchers to precisely study the behaviors of single cells within a defined time period by recording video of them throughout the data collection process. The platform, released in June, works via a microfluidic section with a postage stamp–sized silicon chip containing miniscule NanoPens, long and narrow chambers that isolate and culture individual cells. “If you were the size of a cell, a conference room is about the size of a NanoPen,” says Mark White, senior director of marketing at Berkeley Lights. He declined to name the product’s price, saying that it varies based on applications. “The platform is in the ballpark of a very high-end microscope,” he says. Berkeley Lights’s Lightning optofluidic platform enables researchers to precisely study the behaviors of single cells within a defined time period by recording video of them throughout the data collection process. The platform, released in June, works via a microfluidic section with a postage stamp–sized silicon chip containing miniscule NanoPens, long and narrow chambers that isolate and culture individual cells. “If you were the size of a cell, a conference room is about the size of a NanoPen,” says Mark White, senior director of marketing at Berkeley Lights. He declined to name the product’s price, saying that it varies based on applications. “The platform is in the ballpark of a very high-end microscope,” he says.
Medical oncologist Cassian Yee, a consultant for Berkeley Lights who studies endogenous T cell therapy, uses the device in his lab at the University of Texas MD Anderson Cancer Center. “Every single cell is tracked from beginning to end. . . . This not only allows you to analyze a lot of different parameters at once, even with a few cells, but also if you’re interested in using this as quality control for your T cell product, it’s probably as rigorous as you can get,” he says.
Researchers using other optofluidic devices have to use multiple instruments to perform cell assays and take a snapshot of the cells on each instrument. In addition to consolidating equipment, the Lightning platform runs protocols on Python script, making it access-ible to researchers worldwide. “You can give [a protocol] to somebody else halfway around the world who can run exactly the same protocol,” says Yee. “That’s the beauty of a device like this. . . . This is a sort of quantum leap from what people were doing.”
Cruickshank-Quinn: “Being able to visualize phenotypes and perform functional assays on thousands of individual cells simultan-eously allows users to obtain results in a fraction of the time. Very promising technology for cancer research.”
Pacific Biosciences Sequel II  Pacific Biosciences has multiplied the capacity of its previous DNA- and RNA-sequencing instrument, Sequel, which won a spot in The Scientist’s 2016 Top 10 Innovations. The new Sequel II, priced at $495,000 in the US, can generate about eight times as much data as its predecessor. Inside each of its sample chambers, termed zero-mode waveguides, the instrument detects bases as a polymerase adds them to a nucleotide chain, yielding sequence information. Pacific Biosciences has multiplied the capacity of its previous DNA- and RNA-sequencing instrument, Sequel, which won a spot in The Scientist’s 2016 Top 10 Innovations. The new Sequel II, priced at $495,000 in the US, can generate about eight times as much data as its predecessor. Inside each of its sample chambers, termed zero-mode waveguides, the instrument detects bases as a polymerase adds them to a nucleotide chain, yielding sequence information.
The result, as with the original Sequel, is long-read DNA or RNA sequences, now delivered more quickly and at lower cost, says Marty Badgett, PacBio’s senior director of product management. Long reads allow researchers to identify structural variants in the genome, “which is quite complementary to [information gleaned from] short reads,” such as single nucleotide polymorphisms (SNPs), he says. That structural information includes translocations in the genome, copy number variants, insertions, and deletions.
Because of its long read lengths and flexible run conditions, the Sequel II is adept at de novo genome sequencing in species for which no reference genome is yet available, says Shawn Levy, a geneticist at the nonprofit HudsonAlpha Institute for Biotechnology, which was given early access to the instrument. And, he adds, the instrument’s long-read sequencing is also good for analyzing highly repetitive or homologous regions of the genome. Levy says that he and other researchers at Huntsville, Alabama–based HudsonAlpha are exploring the use of the Sequel II to search for genetic causes of rare diseases for which no SNPs can be found, and to analyze RNA in cancer cells, among other applications.
Wiley: “Does longer reads with greater accuracy, at greater throughput, and at significantly lower cost. PacBio sets the standard for long-read sequencing and this upgrade of their instrument should have high impact on genomics sciences.”
Nanolive CX-A  A live-cell imaging microscope released in July 2019, CX-A promises to answer important questions about cellular interactions “in a much more elegant and powerful way” than ever before, quantitative biologist Mathieu Fréchin tells The Scientist. Fréchin works at Nanolive, the company that developed the microscope, and has used the instrument to observe mitochondrial fission and fusion, the way groups of cells interact and react to one another, and other cellular phenomena. A live-cell imaging microscope released in July 2019, CX-A promises to answer important questions about cellular interactions “in a much more elegant and powerful way” than ever before, quantitative biologist Mathieu Fréchin tells The Scientist. Fréchin works at Nanolive, the company that developed the microscope, and has used the instrument to observe mitochondrial fission and fusion, the way groups of cells interact and react to one another, and other cellular phenomena.
To capture such dynamics, CX-A doesn’t use stains or labels. Instead, it reconstructs a three-dimensional hologram based on how the sample refracts light. To understand how it works, “think of straw in a clear glass of lemonade,” says Alexander Jones, Nanolive’s chief commercial officer. In the air, the straw appears normal, but in the lemonade, it appears bent, or refracted. In a cell, each organelle refracts light differently, as if each organelle were a different lemonade. CX-A accounts for the variations in refraction and reconstructs the three-dimensional image formed by the interference of the refraction of the cellular components in its field of view.
This holographic technique was the backbone of Nanolive 3D Cell Explorer, which won a spot in our 2015 Top 10 Innovations. The company’s new technology builds on the 3D Cell Explorer design and is automated so scientists can program the various fields of view they would like to observe in a single slide or in 96-well plates, then walk away and let the machine do the work. “It’s the difference between the Wright brothers’ airplane and the Concorde,” Jones says. Although Nanolive declined to give a precise cost for CX-A, Jones says that the tool is a tenth of the price of a super-resolution confocal microscope, which typically runs between $300,000 and $500,000.
Kamdar: “Nanolive imaging is a great discovery tool that allows the measurement of cellular processes and kinetics in real-time, enabling multi-parameter analysis at single-cell and subcellular scale.”
Abbott FreeStyle Libre 2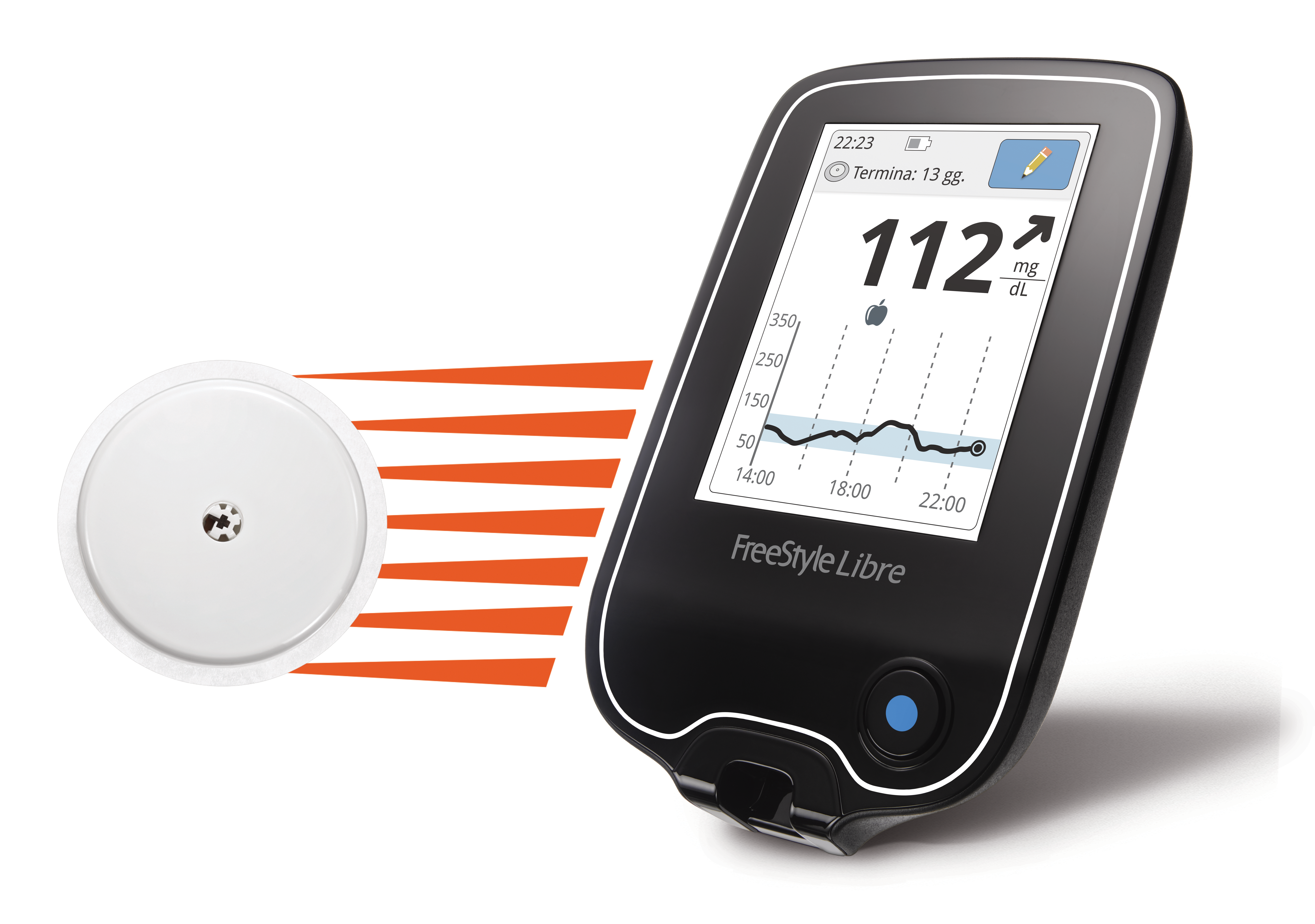  With a swipe of a cellphone across a sensor attached to the back of his arm, Michael Krauser can instantaneously check his glucose levels. For Krauser, a diabetes patient living in Germany, there are no painful finger pricks, and no hassle trying get enough blood on test strips to measure his glucose. Abbott’s FreeStyle Libre 2 is an “elegant” solution to checking his glucose that “adds a certain security to my life,” he says. With a swipe of a cellphone across a sensor attached to the back of his arm, Michael Krauser can instantaneously check his glucose levels. For Krauser, a diabetes patient living in Germany, there are no painful finger pricks, and no hassle trying get enough blood on test strips to measure his glucose. Abbott’s FreeStyle Libre 2 is an “elegant” solution to checking his glucose that “adds a certain security to my life,” he says.
The FreeStyle Libre 2 system was approved for use in Europe in 2018 and is not yet available in the US. When the glucose monitoring system does hit the US market, each sensor, which is temporary and lasts two weeks, will cost about $55. The size of two stacked quarters, a sensor measures glucose through what looks like a pin inserted into the interstitial fluid, a liquid that surrounds the cells just below the skin. Patients can check their glucose by swiping a smartphone loaded with the FreeStyle LibreLink app across the sensor, or they can buy the FreeStyle Libre reader for a one-time cost of $70.
The Bluetooth-enabled sensor allows users to set sound or vibration alarms on their phones or readers to alert them when their blood sugar is too low or too high, so they can bring it back to baseline by eating carbs or injecting insulin. The alerts are extremely helpful to children and other patients who might not other-wise know of extreme changes in their glucose levels, especially during sleep, says Marc Taub, the divisional vice president of product development for diabetes care at Abbott.
“Innovations like the FreeStyle Libre 2 technology give me courage to live a good life with my type 1 diabetes,” Krauser says.
Cruickshank-Quinn: “This offers a great solution for diabetics so that they no longer need constant finger sticks, and the option to use either the reader or smartphone gives patients flexibility of choices, especially in these technologically advanced times.”
Akoya Biosciences CODEX  Until a few years ago, tumor profiling either lacked spatial context, or was limited to just two or three markers at a time, says Julia Kennedy-Darling, director of research and development at Akoya Biosciences. Increasing the amount of spatial information that can be mined from tumors is important, she says, because “we’re understanding that it’s not sufficient just to be able to catalog what cells might be present in a sample, but where they are and which cells are next to each other.” To that end, as a postdoc in Garry Nolan’s lab at Stanford University, Kennedy-Darling codeveloped a technique to detect up to 40 different markers—and indicate their locations—in a single sample. Until a few years ago, tumor profiling either lacked spatial context, or was limited to just two or three markers at a time, says Julia Kennedy-Darling, director of research and development at Akoya Biosciences. Increasing the amount of spatial information that can be mined from tumors is important, she says, because “we’re understanding that it’s not sufficient just to be able to catalog what cells might be present in a sample, but where they are and which cells are next to each other.” To that end, as a postdoc in Garry Nolan’s lab at Stanford University, Kennedy-Darling codeveloped a technique to detect up to 40 different markers—and indicate their locations—in a single sample.
That technique, called CODEX, overcomes the problem of spectral overlap—that is, when too many antibodies fluorescing different colors are added to a sample, researchers can’t reliably distinguish among them using conventional detection methods. The key with CODEX, Kennedy-Darling explains, is that dyes aren’t covalently bonded to particular antibodies, but are induced to associate with different antibodies over multiple cycles of analysis, with three dyes revealed and imaged in each cycle. Akoya declined to reveal CODEX’s price.
The instrument’s capacity to collect data on multiple markers per sample was a selling point for Andras Heczey, an oncology researcher at Baylor College of Medicine who has used CODEX for antibody validation and plans to deploy it to analyze how immunotherapy treatments interact with cancers. “Postinfusion biopsies are extremely precious” because they’re hard to come by, he explains, and they tend to be small. His aim with CODEX is to “get the most data from small samples of tissues to understand as much of the tumor micro-environment as possible.”
Wiley: “Although multiplexed antibody detection systems have been described before, the simplicity . . . of this system should make this powerful approach more generally available.”
Owlstone Medical EVOC Probes  Launched in June, EVOC probes from Owlstone Medical are a new way to measure exogenous volatile organic compounds in the breath. The probes expand upon the company’s Breath Biopsy platform, launched in 2017, in which researchers collect breath samples from patients that are then analyzed in Owlstone Medical’s lab for biomarkers of cancer or other diseases. Prior to the launch of EVOC probes, the company’s breath research was focused on endogenous, or internally generated, biomarkers. EVOC probes, on the other hand, allow scientists to administer small doses of safe volatile organic compounds, such as terpenes, as probes and then measure the concentration of the products of reactions involving those compounds to assess liver function or drug metabolism. “The big advantage is that you know what you’re looking for,” says Billy Boyle, cofounder and CEO of Owlstone Medical. “Rather than having to try and find the tiny needle in the haystack, you’re able to introduce a much larger signal into the system.” Launched in June, EVOC probes from Owlstone Medical are a new way to measure exogenous volatile organic compounds in the breath. The probes expand upon the company’s Breath Biopsy platform, launched in 2017, in which researchers collect breath samples from patients that are then analyzed in Owlstone Medical’s lab for biomarkers of cancer or other diseases. Prior to the launch of EVOC probes, the company’s breath research was focused on endogenous, or internally generated, biomarkers. EVOC probes, on the other hand, allow scientists to administer small doses of safe volatile organic compounds, such as terpenes, as probes and then measure the concentration of the products of reactions involving those compounds to assess liver function or drug metabolism. “The big advantage is that you know what you’re looking for,” says Billy Boyle, cofounder and CEO of Owlstone Medical. “Rather than having to try and find the tiny needle in the haystack, you’re able to introduce a much larger signal into the system.”
The product’s price tag, about $400 per experiment, includes the cost of the probes and the breath collection device and data analysis. The company is currently working with cancer researchers such as Rebecca Fitzgerald at the University of Cambridge to develop tests for clinical settings, where the cost of running breath-based tests would be far cheaper than the current cost in the research sector—on the scale of “tens of dollars,” says Boyle. “The technology is easy to use and generally well tolerated by patients,” Fitzgerald writes in an email to The Scientist. “[Patients] find the process noninvasive compared to other ways of taking samples, such as taking blood samples or tissue biopsies,” adds Irene Debiram-
Beecham, a principal research nurse at the University of Cambridge who helped coordinate recent studies in Fitzgerald’s lab.
Cruickshank-Quinn: “This technology . . . has potential to provide a quick screening tool in the health-care field but to also allow scientists to perform noninvasive studies in a number of research areas, including lung health and disease.”
10X Genomics Chromium Single Cell ATAC Solution  In 2013, researchers at Stanford University reported a new chromatin interrogation method called ATAC-seq (Assay for Transposase Accessible Chromatin). It treats cells with a transposase, an enzyme that cuts open stretches of DNA and adds adapters to those nucleic acids, allowing scientists to then amplify and sequence the fragments (Nat Meth, 10:1213–18, 2013). The group launched a company, Epinomics, to commercialize the technology. Around the same time, 10x Genomics was developing single-cell RNA sequencing using a droplet-based approach, notes Stanford’s Howard Chang, codeveloper of ATAC-seq. “Marrying these two technologies together seemed like a very attractive proposition” that could lead to a high-throughput single-cell ATAC-seq system. In 2013, researchers at Stanford University reported a new chromatin interrogation method called ATAC-seq (Assay for Transposase Accessible Chromatin). It treats cells with a transposase, an enzyme that cuts open stretches of DNA and adds adapters to those nucleic acids, allowing scientists to then amplify and sequence the fragments (Nat Meth, 10:1213–18, 2013). The group launched a company, Epinomics, to commercialize the technology. Around the same time, 10x Genomics was developing single-cell RNA sequencing using a droplet-based approach, notes Stanford’s Howard Chang, codeveloper of ATAC-seq. “Marrying these two technologies together seemed like a very attractive proposition” that could lead to a high-throughput single-cell ATAC-seq system.
10X acquired Epinomics in late August of last year, and a little more than a month later, launched the Chromium Single Cell ATAC Solution, at a cost of $1,500 per sample. The product partitions individual cell nuclei into droplets and uses genetic barcodes to tag relevant sequences—in this case, those in the open chromatin position. The DNA can then be sequenced en masse and sorted bioinformatically to determine which fragments came from which cells. 10X’s droplet-based approach scaled the throughput of single-cell ATAC sequencing from a few hundred that could be done manually to thousands or tens of thousands of cells per run, says Fergus Chan, cofounder of Epinomics and now director of product management at 10X.
Immunologist Ansuman Satpathy, a former postdoc in Chang’s group, now uses the Chromium Single Cell ATAC Solution in his own lab at Stanford to study immune cell biology in the context of cancer. Satpathy notes that the technology yields data on many different levels—from individual enhancers to the genes that they regulate to genome-wide transcription factor activity. “It gives you [insights into] many more features of a cell than you could get from other assays,” Satpathy says.
Kamdar: “Discover cellular heterogeneity stemming from epigenetic variability. Accelerates the understanding of the regulatory landscape of the genome, thereby providing insights into cell variability.”
Horizon Discovery Edit-R all-in-one lentiviral sgRNA  Horizon Discovery is back among the Top 10 Innovations this year with another CRISPR product—the Edit-R all-in-one lentiviral sgRNA. The company won a spot in last year’s Top 10 with an mRNA that codes for the DNA-cutting nuclease Cas9, while this year’s winning invention combines the sequences for Cas9 and the single guide RNA (sgRNA) that leads the enzyme to the appropriate spot in the genome, all packaged into a viral vector. Horizon Discovery is back among the Top 10 Innovations this year with another CRISPR product—the Edit-R all-in-one lentiviral sgRNA. The company won a spot in last year’s Top 10 with an mRNA that codes for the DNA-cutting nuclease Cas9, while this year’s winning invention combines the sequences for Cas9 and the single guide RNA (sgRNA) that leads the enzyme to the appropriate spot in the genome, all packaged into a viral vector.
Horizon offers predesigned guide RNAs to knock out any gene in the human, mouse, or rat genomes. If users work on any one of 40 other species, they can get custom-made reagents. “Enter in the species, the gene of interest, where you want the guide to edit, and we package it into the all-in-one vector,” says Ryan Donnelly, the product manager for the gene editing portfolio at Horizon. He says the company designed the algorithm that builds the guide RNAs not just to make cuts efficiently, but to reliably knock out specific genes in doing so.
Since the product came out in November 2018, Donnelly says, customers have primarily used it to validate a few genes of interest from larger functional screens or for quick proof-of-principle tests to check if a gene is involved in a particular phenotype they’re studying. (The company did not provide a user to comment by deadline.) Prices range from $500 to $1,000, depending on whether scientists select plasmids or ready-to-use viral particles and off-the-shelf or bespoke guide RNAs.
Wiley: “You pick the gene of interest, and they supply the virus particles that will knock it out at a very reasonable price. Only a single selection step is required. Definitely could greatly accelerate research in understanding gene function in cells.”
|


No comments